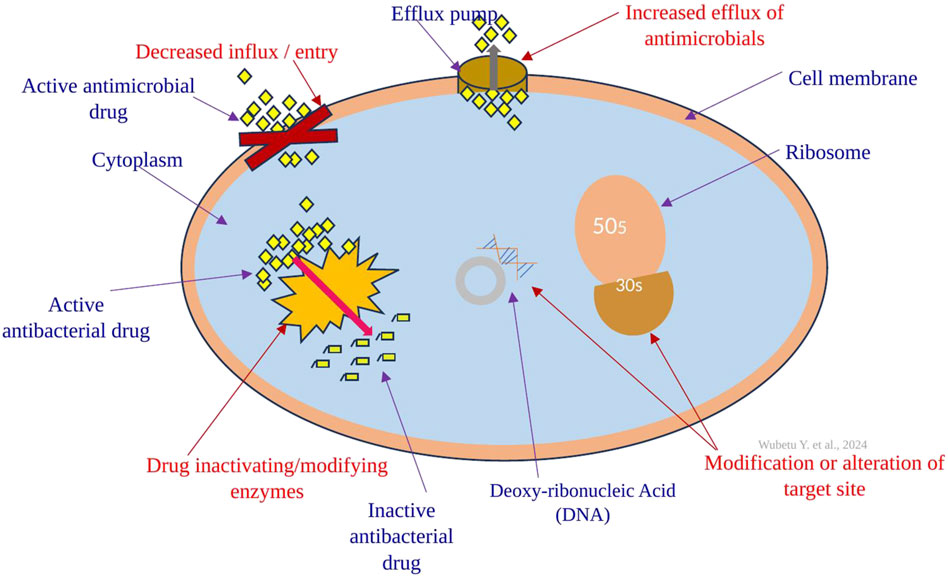
Report on Antimicrobial Resistance in Overlooked Environmental Enterobacterales and Implications for Sustainable Development Goals
Executive Summary
This report details a study on antimicrobial resistance (AMR) and biofilm formation in 14 environmental isolates of six neglected Enterobacterales species. The findings reveal significant levels of multidrug resistance (MDR) and the presence of numerous antibiotic resistance genes (ARGs) in bacteria from aquatic environments. These results underscore a critical challenge to several Sustainable Development Goals (SDGs), particularly SDG 3 (Good Health and Well-being) and SDG 6 (Clean Water and Sanitation). The dissemination of AMR through water systems represents a direct threat to public health and environmental stability, necessitating integrated surveillance and management strategies aligned with the One Health approach and the 2030 Agenda for Sustainable Development.
1.0 Introduction: The Confluence of AMR and Sustainable Development
The rise of antimicrobial resistance (AMR) is a global crisis that undermines modern medicine and threatens public health. This issue is intrinsically linked to the United Nations Sustainable Development Goals. The dissemination of antibiotic-resistant bacteria and their genetic determinants through environmental pathways, particularly aquatic ecosystems, directly compromises progress toward:
- SDG 3 (Good Health and Well-being): By creating reservoirs of pathogens that can cause difficult-to-treat infections.
- SDG 6 (Clean Water and Sanitation): By highlighting the failure of current water and wastewater management systems to contain biological hazards like AMR.
This study focuses on frequently overlooked species within the order Enterobacterales, which are often not included in standard AMR surveillance but may act as significant environmental hosts of resistance. By analyzing isolates from surface water, groundwater, and wastewater, this investigation provides critical data on the environmental dimension of AMR, a key component of the One Health framework.
2.0 Analysis of Environmental Isolates
Fourteen environmental isolates were analyzed, belonging to six species: Cronobacter sakazakii, Kluyvera intermedia, Leclercia adecarboxylata, Raoultella ornithinolytica, Raoultella terrigena, and Yersinia massiliensis. These were recovered from various water sources, representing potential points of human and environmental exposure.
2.1 Phenotypic and Genotypic Resistance Profiles
The analysis revealed a concerning prevalence of resistance, directly impacting SDG 3 by contributing to the pool of environmental AMR.
- Phenotypic Resistance:
- Ten of the fourteen isolates (71%) were classified as multidrug-resistant (MDR).
- High levels of resistance were observed against ampicillin and tetracycline.
- Intrinsic resistance to erythromycin was widespread.
- Notably, no isolates exhibited resistance to carbapenems, a last-resort class of antibiotics, although one strain showed intermediate resistance.
- Genotypic Resistance (ARG Screening):
- A strong correlation was found between phenotypic resistance and the presence of corresponding ARGs.
- Thirteen different ARGs were detected across the isolates.
- The most prevalent genes were β-lactamases (blaTEM, blaCTX-M), which confer resistance to common penicillins and cephalosporins.
- Genes conferring resistance to tetracyclines (tet), sulfonamides (sul), macrolides (erm, mef), and quinolones (qnr) were also identified.
- The class 1 integron-integrase gene (intI1), a marker for mobile genetic elements that facilitate the spread of AMR, was detected in one isolate.
2.2 Biofilm Formation and Genetic Diversity
Biofilm formation and genetic plasticity are key survival strategies for bacteria, influencing their persistence in environments and their potential to spread AMR, thereby affecting both SDG 6 and SDG 14 (Life Below Water).
- Biofilm Production: Half of the isolates were capable of weak biofilm formation. A positive correlation was observed between biofilm-forming ability and the MDR phenotype, suggesting that this survival mechanism may enhance persistence under antibiotic pressure.
- Genetic Fingerprinting (ERIC-PCR): The analysis revealed significant genetic diversity among the isolates. An important negative correlation was found between the number of ERIC bands (a marker of genome plasticity) and the levels of both phenotypic and genotypic resistance. This suggests that strains with higher genomic plasticity may rely less on specific resistance mechanisms, a finding that warrants further investigation.
3.0 Implications for Sustainable Development Goals (SDGs)
The findings of this study have direct and significant implications for the achievement of several SDGs.
3.1 SDG 3: Good Health and Well-being
The presence of MDR pathogens like Cronobacter sakazakii, Kluyvera intermedia, and Raoultella ornithinolytica in environmental water sources constitutes a direct public health risk. These “overlooked” bacteria serve as environmental reservoirs of ARGs, which can be transferred to human pathogens, complicating treatments and increasing morbidity and mortality from infections.
3.2 SDG 6: Clean Water and Sanitation
The study highlights that aquatic environments, including surface water, groundwater, and especially wastewater, are critical vectors for the dissemination of AMR. The recovery of resistant bacteria from treated wastewater effluent indicates that current sanitation and water treatment processes are inadequate for mitigating this biological threat. Achieving Target 6.3 (improve water quality by reducing pollution, halving the proportion of untreated wastewater, and substantially increasing recycling) is crucial for controlling the environmental spread of AMR.
3.3 SDG 11: Sustainable Cities and Communities
Urban water cycles, which collect residential, hospital, and industrial runoff, concentrate antimicrobial residues and resistant bacteria in wastewater treatment plants. These facilities can become hotspots for the evolution and dissemination of AMR. Ensuring sustainable and safe urban water management is essential for protecting community health from environmental pathogens.
3.4 SDG 14 & 15: Life Below Water and Life on Land
The release of antibiotic-resistant bacteria into natural ecosystems disrupts the native microbiome and threatens biodiversity. The spread of ARGs can have unforeseen ecological consequences, altering microbial community structures and functions in both aquatic and terrestrial environments.
4.0 Conclusion and Recommendations
This report confirms that overlooked environmental bacteria in aquatic ecosystems are significant hosts of antimicrobial resistance, posing a multifaceted threat to public health and environmental integrity. The findings directly challenge the progress towards achieving key Sustainable Development Goals, particularly SDG 3 and SDG 6.
Based on these findings, the following recommendations are proposed:
- Expand AMR Surveillance: Monitoring programs must be expanded to include environmental compartments (water, soil) and a broader range of bacterial species beyond common clinical pathogens. This aligns with a One Health approach to AMR.
- Improve Water and Wastewater Treatment: Investment in advanced water treatment technologies is necessary to remove or inactivate resistant bacteria and ARGs before effluent is discharged into the environment, directly supporting SDG 6.
- Integrate AMR into Environmental Policy: AMR should be recognized as a critical pollutant. Policies governing water quality and environmental protection must incorporate standards and targets for reducing AMR dissemination.
- Promote Further Research: Further investigation is needed to understand the dynamics of AMR in the environment, including the role of genome plasticity in bacterial survival and the ecological impact of AMR pollution on achieving SDG 14 and SDG 15.
Addressing the environmental dimension of AMR is not merely a technical challenge but a fundamental requirement for sustainable development and global health security.
1. Which SDGs are addressed or connected to the issues highlighted in the article?
The article primarily addresses issues related to two Sustainable Development Goals:
-
SDG 3: Good Health and Well-being
The article’s core focus is on antimicrobial resistance (AMR), which it describes as a “critical health concern” and an “antibiotic resistance crisis.” It investigates emerging and opportunistic pathogens like Cronobacter sakazakii, which are reported to cause “rare infections, severe illnesses, and deaths.” This directly connects to ensuring healthy lives and promoting well-being by combating communicable diseases and managing global health risks.
-
SDG 6: Clean Water and Sanitation
The study analyzes bacterial isolates from various aquatic environments, including “surface water, groundwater and wastewater samples.” It explicitly links the spread of pathogens and AMR to “faecal pollution” and “water contamination.” The research on bacteria in wastewater influent and effluent directly pertains to water quality, pollution reduction, and wastewater treatment, which are central themes of SDG 6.
2. What specific targets under those SDGs can be identified based on the article’s content?
Based on the issues discussed, the following specific targets are relevant:
-
SDG 3: Good Health and Well-being
-
Target 3.3: By 2030, end the epidemics of AIDS, tuberculosis, malaria and neglected tropical diseases and combat hepatitis, water-borne diseases and other communicable diseases.
The article supports this target by investigating emerging pathogens that cause communicable and water-borne diseases. It notes that “most of the major emerging infectious diseases caused by bacterial agents originated either from an animal, as zoonoses, or from contaminated water sources.” The study of bacteria like Cronobacter sakazakii, linked to “life-threatening infections in neonates,” and Leclercia adecarboxylata, “often acquired via wound contact with an aquatic environment,” directly relates to understanding and combating such diseases.
-
Target 3.d: Strengthen the capacity of all countries… for early warning, risk reduction and management of national and global health risks.
Antimicrobial resistance is a recognized global health risk. The article contributes to this target by conducting “antimicrobial surveillance in the environment” to assess the “occurrence and health risks represented by rare and overlooked bacteria.” By identifying AMR profiles and resistance genes (ARGs), the study provides data crucial for early warning systems and risk management strategies to mitigate the spread of resistant pathogens.
-
-
SDG 6: Clean Water and Sanitation
-
Target 6.1: By 2030, achieve universal and equitable access to safe and affordable drinking water for all.
The article connects to this target by discussing water quality assessment. It mentions that Escherichia coli is “the most widely used faecal indicator in drinking water risk management.” The study’s investigation into other “overlooked environmental Enterobacterales” in various water sources, including groundwater, highlights the complexity of ensuring water is free from harmful pathogens, a prerequisite for it to be considered “safe.”
-
Target 6.3: By 2030, improve water quality by reducing pollution, eliminating dumping and minimizing release of hazardous chemicals and materials, halving the proportion of untreated wastewater…
This target is directly addressed through the article’s focus on wastewater as a reservoir for antibiotic-resistant bacteria. The study analyzes isolates from both wastewater influent and effluent, which is a method to assess the effectiveness of wastewater treatment in removing pathogens. The finding that “anthropogenic-impacted environments are affected by the release of pathogens and antibiotic resistant bacteria” underscores the need to reduce pollution from untreated wastewater to protect public health and the environment.
-
3. Are there any indicators mentioned or implied in the article that can be used to measure progress towards the identified targets?
The article mentions or implies several indicators that can be used to measure progress:
-
Indicators for SDG 3 Targets
-
For Target 3.3 (Combat communicable diseases):
An implied indicator is the incidence and prevalence of infections caused by emerging and multidrug-resistant pathogens in different environments. The article’s focus on identifying and characterizing bacteria like Cronobacter sakazakii, Kluyvera intermedia, and Raoultella ornithinolytica provides the foundational data needed to track these specific water-related health threats.
-
For Target 3.d (Manage health risks):
The article uses direct indicators to measure the risk of AMR:
- Proportion of bacteria resistant to specific antibiotics: The study measures phenotypic resistance, noting that isolates showed “increased resistance to ampicillin and tetracycline.”
- Prevalence of multidrug-resistant (MDR) bacteria: The article quantifies this directly, stating, “Ten isolates were MDR.”
- Presence and frequency of specific antibiotic resistance genes (ARGs): The study screens for 24 ARGs and reports the detection of genes like blaTEM, blaCTX-M, tet, and sul, which serve as genotypic indicators of resistance potential.
-
-
Indicators for SDG 6 Targets
-
For Target 6.1 (Safe drinking water):
The article references the use of faecal indicator bacteria (e.g., E. coli) in water quality assessment. This is a key component of the official indicator 6.1.1 (Proportion of population using safely managed drinking water services), as “safely managed” implies freedom from faecal contamination. The study suggests the need for broader indicators by examining other pathogenic Enterobacterales found in water sources.
-
For Target 6.3 (Improve water quality/wastewater treatment):
The study’s methodology implies indicators relevant to both official indicators under this target:
- Assessment of wastewater treatment efficacy (related to Indicator 6.3.1): By isolating bacteria from both “wastewater influent and effluent,” the study provides a method to measure the reduction of pathogenic and resistant bacteria, which is a measure of how “safely treated” the wastewater is.
- Ambient water quality (related to Indicator 6.3.2): The presence and concentration of antibiotic-resistant bacteria and their genes in “surface water” and “groundwater” serve as a direct indicator of water body pollution and quality, especially in “anthropogenic-impacted environments.”
-
4. Table of SDGs, Targets, and Indicators
| SDGs | Targets | Indicators Identified in the Article |
|---|---|---|
| SDG 3: Good Health and Well-being | 3.3: End epidemics and combat water-borne and other communicable diseases. |
|
| 3.d: Strengthen capacity for early warning, risk reduction, and management of global health risks. |
|
|
| SDG 6: Clean Water and Sanitation | 6.1: Achieve access to safe and affordable drinking water. |
|
| 6.3: Improve water quality by reducing pollution and increasing wastewater treatment. |
|
Source: nature.com